METHODS
Membrane proteins are some of the most important proteins in biology but they are also some of the most difficult proteins to study. Due to their hydrophobic nature, these proteins tend to aggregate and precipitate in aqueous solutions. For this reason these proteins must be incorporated into detergents or lipids that resemble their native environment. As a result, expression and purification of these proteins can be a daunting task. The methods used in our lab allow us to produce milligram amounts of membrane proteins for our studies.
Expression and Purification of Membrane Proteins
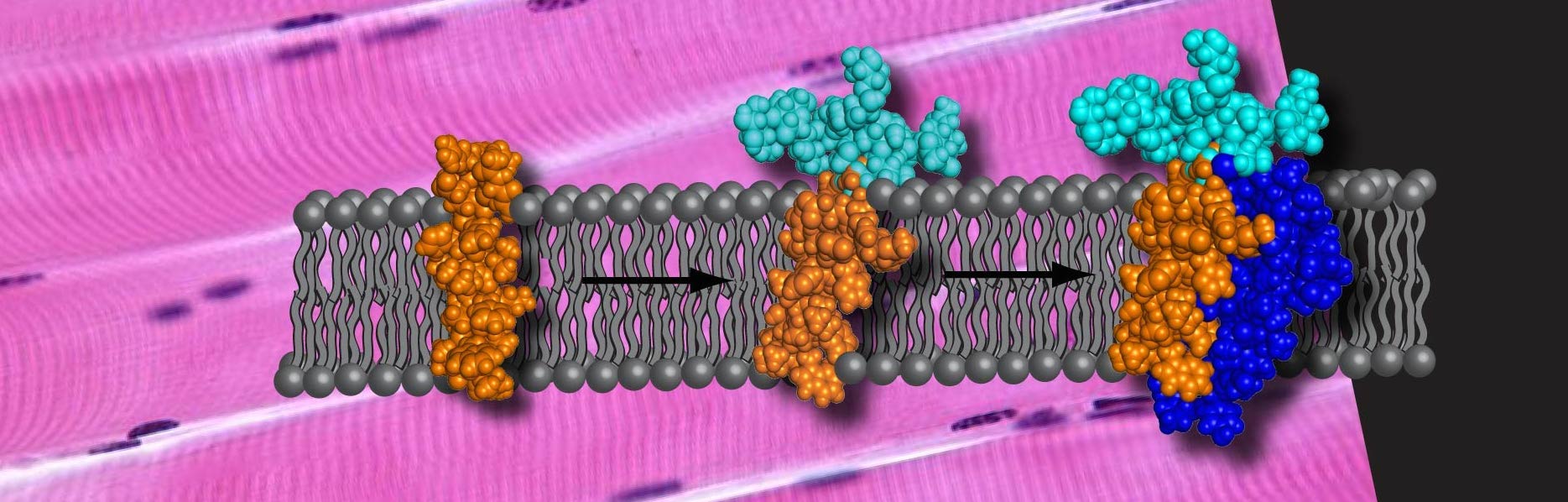
Membrane proteins are incredibly difficult to study due to their hydrophobic nature. Unfortunately, our research requires us to produce milligram amounts of protein for our samples. Recombinant expression of full-length membrane proteins are often avoided due to their lethality to the bacteria during over-expression. Once the protein is expressed it is tremendously difficult to purify the protein due to its propensity to aggregate, even in the presence of detergents or lipids.
Lipid Assemblies
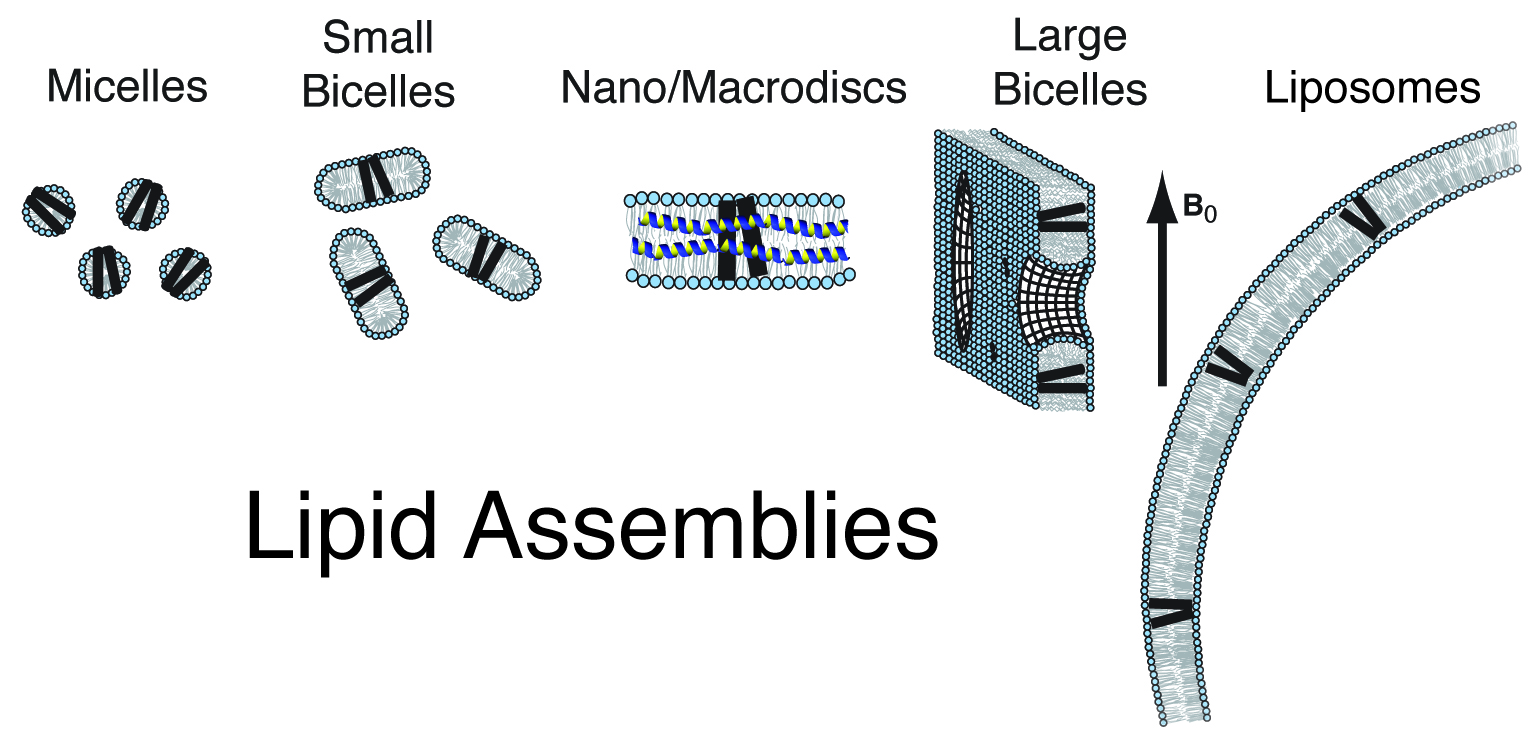
A wide range of detergent and lipid environments have been used for looking at membrane proteins by NMR spectroscopy. By using nanodisks and macrodisks we are able to look at membrane proteins by both solution and solid-state NMR in a membrane-like environment. In order to study the structure, dynamics and function of this protein it will be incorporated into lipid nanodiscs. These lipid assemblies provide the ideal environment for studying these proteins in a bilayer environment. Protein/nanodisc samples will be made into samples for solution and solid-state NMR.
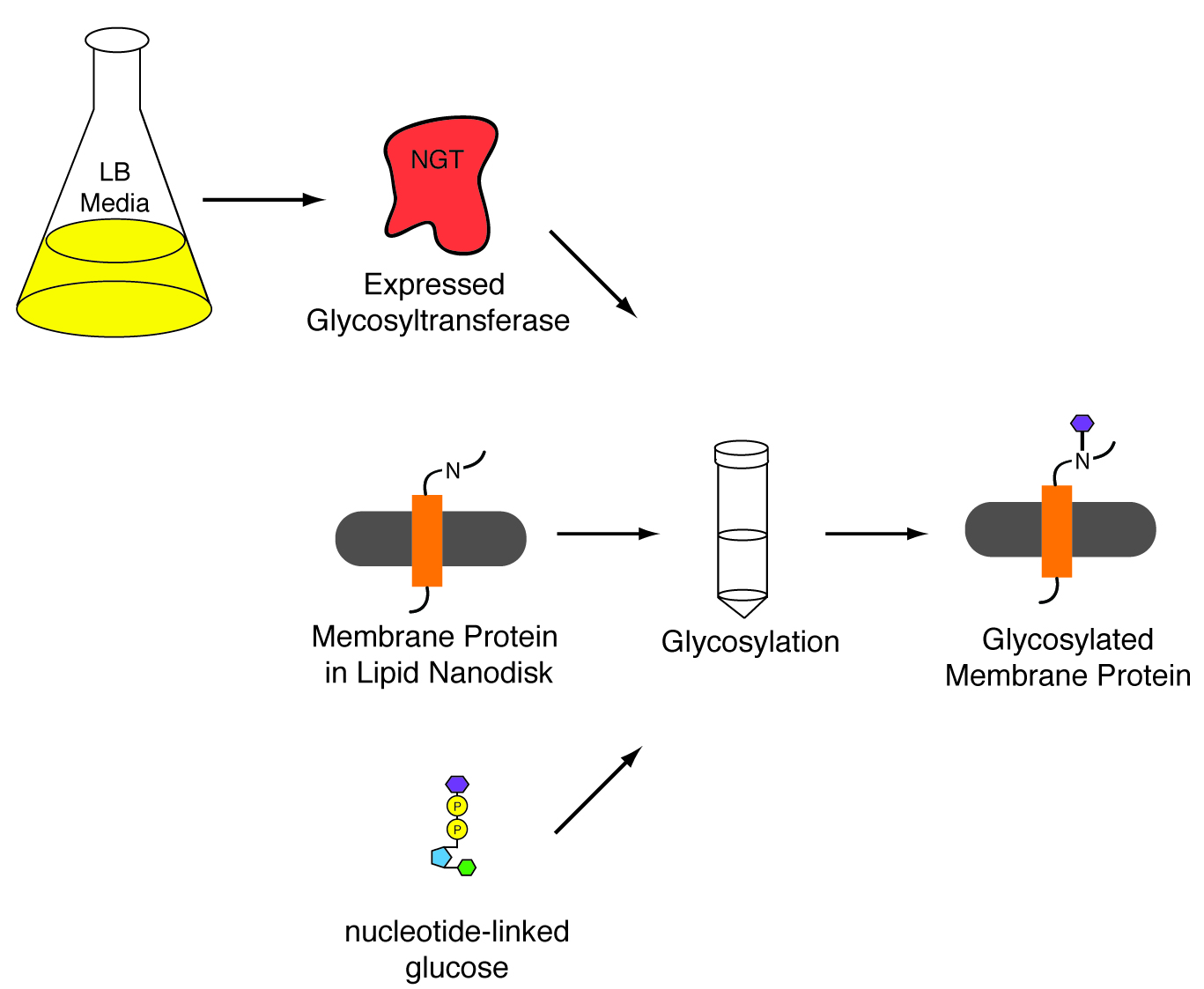
In vitro glycosylation
In order to study our membrane proteins with a saccharide attached we we have to find a way to glycosylate our proteins. Using methods first described by Aebi and co-workers* we will attempt to glycosylate our membrane protein in nanodisks. The nanodisk provides a membrane-like environment that allows the native structure of our membrane protein to be maintained while providing a detergent free system for the glucotransferase to function properly. (*Schwarz, F. et al, JBiolChem, 2011, 286:35267-74)
NMR
Once these proteins are purified and glycosylated we use Nuclear Magnetic Resonance (NMR) Spectroscopy to determine the structure, dynamics and interactions of these proteins. The method measures the chemical environment around the nuclei of atoms that make up the protein. We can measure bond angles, intramolecular distances and intermolecular distances to develop a 3D model of the protein.
Interaction Studies
The dipolar coupling measurements that acquired can be converted into angle constraints for the structure calculation of our protein structures. A high-resolution 3D structure can be determined using these methods. Other measurements can also be performed including protein-protein and drug-protein interactions. By adding the interacting molecule and measuring the chemical shift perturbations and line-shape changes we are able to determine the precise locations of those interactions. These measurements can be done in the presence and absence of the saccharide to determine the effects that the glycosylation has on these properties as well.